| Patient Information: An 8-year-old Caucasian girl |
| Disease: Mental Retardation Syndrome ( DOID : 1059 ) All events related to this disease. : 1059 ) All events related to this disease.
|
|
| Precision: C |
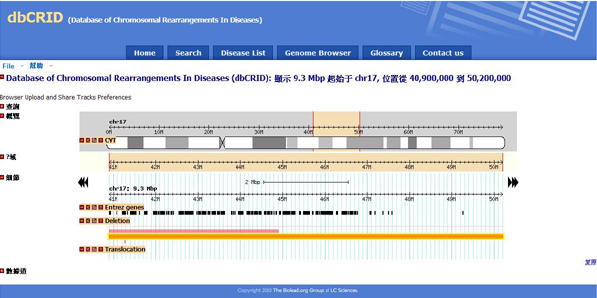
| Chromosome Rearrangement Type: Deletion |
| Karyotype : del(16) (p13.3p13.3) : del(16) (p13.3p13.3) |
| First Breakpoint : : |
| Second Breakpoint : : |
| Length of Deletion Segment is 1Mb. |
| Junction Sequence is Unavailable . . |
| Experimental Method: FISH, CGH |
| PubMed ID: 18076105 |
| Authors: Gibson WT,Harvard C,Qiao Y,Somerville MJ,Lewis
ME,Rajcan-Separovic E |
| Journal: American journal of medical genetics. Part
A |
 : 10789 ( Case ID
: 10789 ( Case ID : 11635 )
: 11635 ) : 1059 ) All events related to this disease.
: 1059 ) All events related to this disease.
 dbCRID
dbCRID