Event ID : 10047 ( Case ID : 10047 ( Case ID : 11923 ) : 11923 ) |
Disease: Chronic Myeloid Leukemia ( DOID : 8552 ) All events related to this disease. : 8552 ) All events related to this disease.
|
||||
| Event Information |
Precision: B |
|||||
Chromosome Rearrangement Type: Insertion |
|||||
| Chromosome: 22 | |||
Karyotype : ins(22;9) (q11.2;q34.1q34.2) : ins(22;9) (q11.2;q34.1q34.2) | |||
| Breakpoints Information: | ||||
Breakpoint : : | |||
| Location: q11.23 | |||
| This breakpoint occurs in NM_004327.3. | |||
| Insertion Source is chromosome 9 q34.12q34.22. | |||
#Breakpoint is Unavailable | ||||
| Experimental Method: FISH | ||||
| Reference Information |
| PubMed ID: 18638369 | ||||
| Authors: Virgili A,Brazma D,Reid AG,Howard-Reeves J,Valgañón M,Chanalaris A,De Melo VA,Marin D,Apperley JF,Grace C,Nacheva EP | ||||
| Journal: Molecular Cytogenetics | ||||
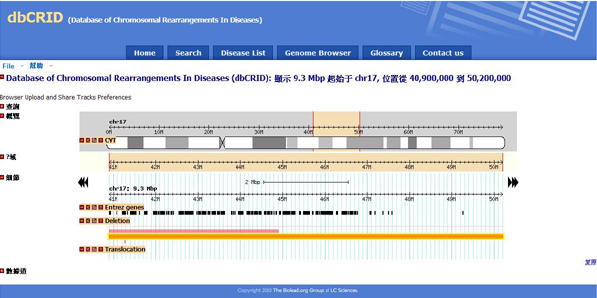
| Go to Genome Browser ( Click the image ) |
 |
|||||
 dbCRID
dbCRID